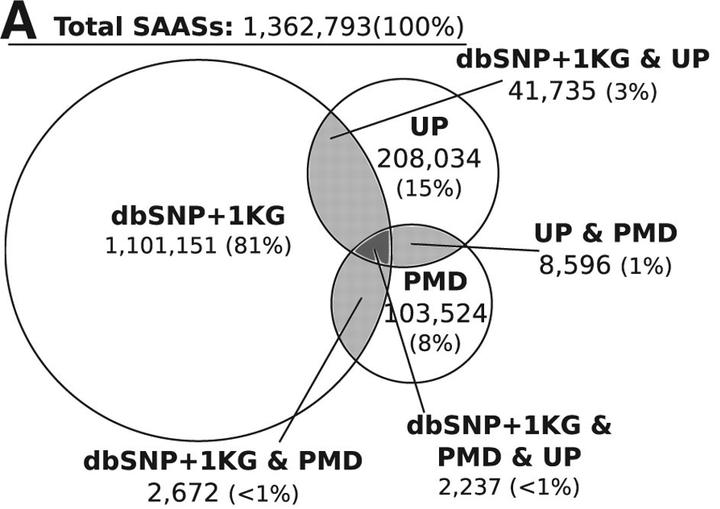
 Venn diagrams describing the overlap of all SNPdbe component database
Venn diagrams describing the overlap of all SNPdbe component database
SNPdbe is a database of single amino acid substitutions and their predicted and experimentally derived functional effects. It was developed by Christian Schaefer at the Rost Lab in Technical University of Munich (TUM) under the supervision of Dr.Burkhard Rost and Dr. Yana Bromberg Most single amino acid substitutions (SAASs) lack experimental annotation of their functional impact. SNPdbe is a database and a webinterface that is designed to fill the annotation gap left by the high cost of experimental testing for functional significance of protein variants. It joins related bits of knowledge, currently distributed throughout various databases, into a consistent, easily accessible, and updatable resource. We currently cover over 155,000 protein sequences from more than 2,600 organisms. Overall we reference more than one million SAASs consisting of natural variants (1000 Genomes data, dbSNP, etc) and SAASs from mutagenesis experiments (SwissProt, PMD, etc). We also report sequence mismatches resulting from differing sequence reports for the same gene. For each SNPdbe entry all available experimental and prediction data is reported and available for download with a click of a button.
If you find SNPdbe useful please cite us!