 Box-plot-and-instance representation of percentages of pEffect-predicted type III effectors
Box-plot-and-instance representation of percentages of pEffect-predicted type III effectors
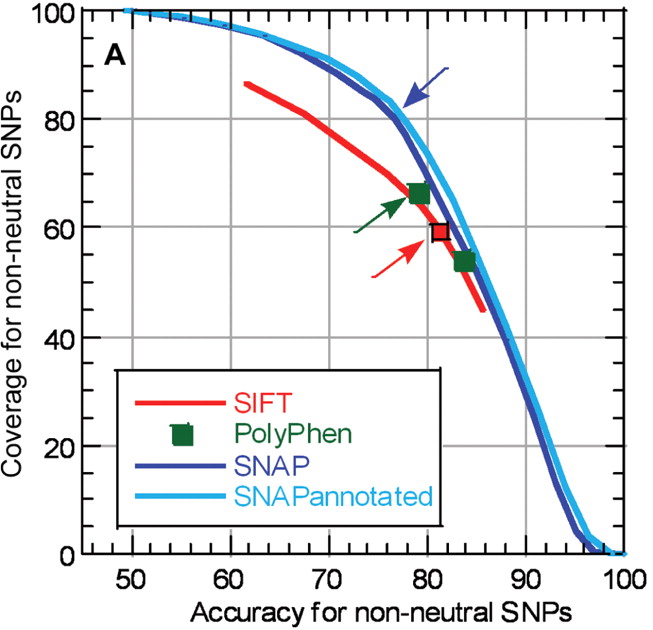
SNAP is a method for evaluating effects of single amino acid substitutions on protein function. It was developed by Yana Bromberg at the Rost Lab, at Columbia University, New York. Single Nucleotide Polymorphisms (SNPs) represent a very large portion of all genetic variations. SNPs found in the coding regions of genes are often non-synonymous, changing a single amino acid in the encoded protein sequence. SNPs are either “neutral” in the sense that the resulting point-mutated protein is not functionally discernible from the wild-type, or they are “non-neutral” in that the mutant and wild-type differ in function. The ability to identify non-neutral substitutions in an ocean of SNPs could significantly aid targeting disease causing detrimental mutations, as well as SNPs that increase the fitness of particular phenotypes. SNAP is a neural-network based method that uses in silico derived protein information (e.g. secondary structure, conservation, solvent accessibility, etc.) in order to make predictions regarding functionality of mutated proteins. The network takes protein sequences and lists of mutants as input, returning a score for each substitution. These scores can then be translated into binary predictions of effect (present/absent) and reliability indices (RI). SNAP returns its results to the user via e-mail.
If you find SNAP useful please cite us!