 Predictprotein webservice (https://open.predictprotein.org)
Predictprotein webservice (https://open.predictprotein.org)
A docker image for the predictprotein (RostLab) pipeline.
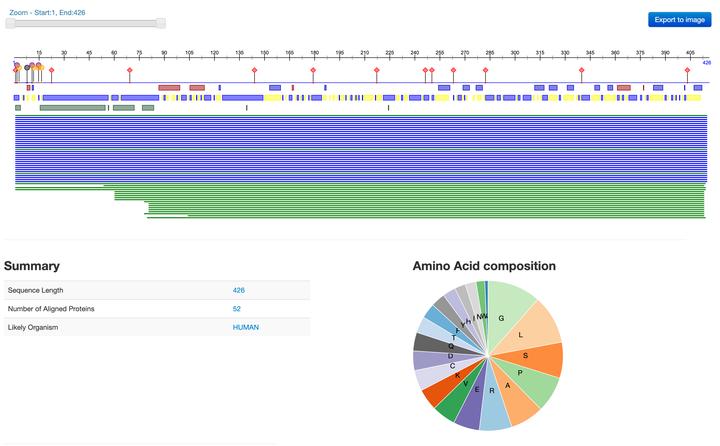
PredictProtein started out by predicting secondary structure and returning families of related proteins. Solvent accessibility and transmembrane helix prediction followed suit shortly thereafter. Over the two decades that PredictProtein has been operating, we have substantially expanded the breadth of structural annotations, e.g. by adding predictions of non-regular secondary structure and intrinsically disordered regions, disulphide bridges and inter-residue contacts, and finally by also covering trans-membrane beta barrels structures. We have also added important resources for the prediction of protein function, e.g. assisting in the annotation of subcellular localization (LocTree, LocTree2, NLSpred), identifying protein-protein interaction sites (ISIS) and protein-DNA binding sites (DISIS & SomeNA to be released shortly). We have added a few simple tools to predict enzymatic activity (unfortunately, this method did not survive the move from New York to Munich) and to infer GeneOntology numbers through sequence homology (MetaStudent). Another major addition are the tools that predict the effect of amino acid changes upon protein function (SNAP & SNAP2), and directly upon protein structure (in preparation).