New publication by Yannick Mahlich and others
 New publication by Yannick Mahlich and others
New publication by Yannick Mahlich and others
HFSP: high speed homology-driven function annotation of proteins Yannick Mahlich, Martin Steinegger, Burkhard Rost, Yana Bromberg
Bioinformatics, Volume 34, Issue 13, 1 July 2018, Pages i304–i312 https://doi.org/10.1093/bioinformatics/bty262
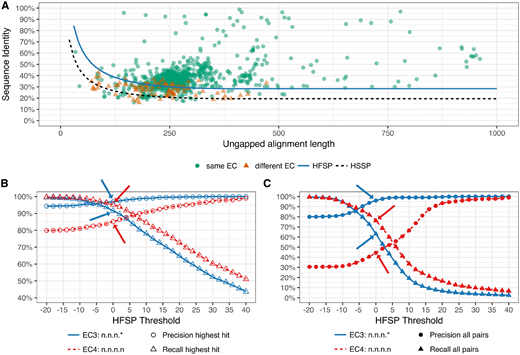
The rapid drop in sequencing costs has produced many more (predicted) protein sequences than can feasibly be functionally annotated with wet-lab experiments. Thus, many computational methods have been developed for this purpose. Most of these methods employ homology-based inference, approximated via sequence alignments, to transfer functional annotations between proteins. The increase in the number of available sequences, however, has drastically increased the search space, thus significantly slowing down alignment methods.Here we describe homology-derived functional similarity of proteins (HFSP), a novel computational method that uses results of a high-speed alignment algorithm, MMseqs2, to infer functional similarity of proteins on the basis of their alignment length and sequence identity. We show that our method is accurate (85% precision) and fast (more than 40-fold speed increase over state-of-the-art). HFSP can help correct at least a 16% error in legacy curations, even for a resource of as high quality as Swiss-Prot. These findings suggest HFSP as an ideal resource for large-scale functional annotation efforts.