Bromberg Lab
Decoding the blueprint of Life
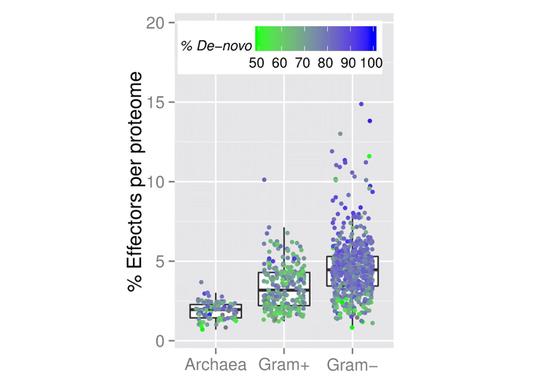
Modern biology increasingly relies on high-throughput techniques. This trend challenges computational biologists to quickly extract as much useful information from the data as possible. In the genomic sense, this primarily implies correlating phenotypic differences with observed nucleotide sequence variations. On the protein side the challenge generally is to annotate protein function at reasonable accuracy levels. The whole organism level, then incorporates all types of evidence to annotate evolutionary history, current health conditions, and prognosed phenotypic changes.
We believe that nucleic and amino acid sequences contain a large portion of the information necessary to address both of these directions. However, we are always willing to supplement this data with other sources available for computational access. The main interest of this lab is in developing fast, accurate, and meaningful ways of analyzing the growing deluge of biological data and in bringing these developments bench- (or patient-) side. To make our predictions we rely on a number of sequence-based features (including evolutionary information, predicted structural features, and available annotations) and utilize a variety of computational methodologies (including artificial learning, network analysis and statistical methods).
Active projects and services in the lab can be explored here.
We are always looking for interested/qualified individuals to join our team!
Jobs
We have open positions!
Two post-doctoral positions are immediately available in the lab of Dr. Yana Bromberg, in the departments of Biology and Computer Science, Emory University, Atlanta (possibly joint with the Institute of Advanced Studies, Technical University of Munich).
We are seeking highly motivated scholars to continue training in an exciting research laboratory at Emory with a focus on molecular functionality encoded in genome and metagenome data. The Bromberg lab is purely computational, studying interactions between the host and the microbiome in light of health and disease. We are also exploring biotic molecular functionality at the origins of life.
Applicants must hold a Ph.D. in Computational Biology, Bioinformatics, or related fields. Programming skills are essential, as well as some familiarity with the major bioinformatics tools/databases. Experience with high performance computing, machine learning, and whole genome and metagenome analysis is highly desired, but not required.
Interested persons should e-mail a cover letter and C.V. to Dr. Yana Bromberg at yana@bromberglab.org. Please visit http://bromberglab.org for more information
updated: 09/2022
Featured
BrombergLab in the News
Did the Seeds of Life Ride to Earth Inside an Asteroid?

Link to publication: DOI: 10.1126/sciadv.abj3984
New study sheds light on origins of life on Earth

Link to publication: DOI: 10.1126/sciadv.abj3984
Genome Analysis Now Allows Scientists To Predict if You Will Have a Miscarriage
Link to publication: DOI: 10.1007/s00439-022-02450-z
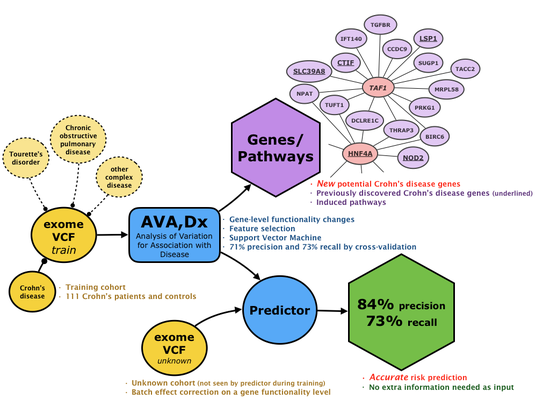
New AI Method May Boost Crohn’s Disease Insight and Improve Treatment

Link to publication: DOI: 10.1186/s13073-019-0670-6
Yana Bromberg
Principal Investigator - Professor of Bioinformatics
BrombergLab
Department of Biology
Department of Computer Science
Emory University
Principal Investigator
Dr. Yana Bromberg’s research focuses on deciphering the DNA “blueprints” of life’s molecular machinery. She develops novel bioinformatics techniques to find out where this machinery comes from and why/how it runs. The answers to these basic questions are important for improving our health/quality of life, preserving our environment, and, well… did we really start as green slime?!
Dr. Bromberg received her degrees from SUNY Stony Brook and Columbia University. Her work has been recognized by private and federal agencies, including NASA and NIH. She received an NSF CAREER award and is also a Fellow of the Munich Institute for Advanced Study. Her findings consistently indicate that our world functions via dependencies and interactions at all scales.
- Protein Function and Variant Effect Prediction
- Artificial Intelligence
-
Ph.D. in Biomedical Informatics (Bioinformatics Track), 2007
Columbia University, New York, NY
-
M.Phil. in Biomedical Informatics, 2004
Columbia University, New York, NY
-
B.A. Biology / B.Eng. Computer Science (Magna Cum Laude), 2001
State University of NY (SUNY) at Stony Brook, Stony Brook, NY
Projects
Selected projects & web-services
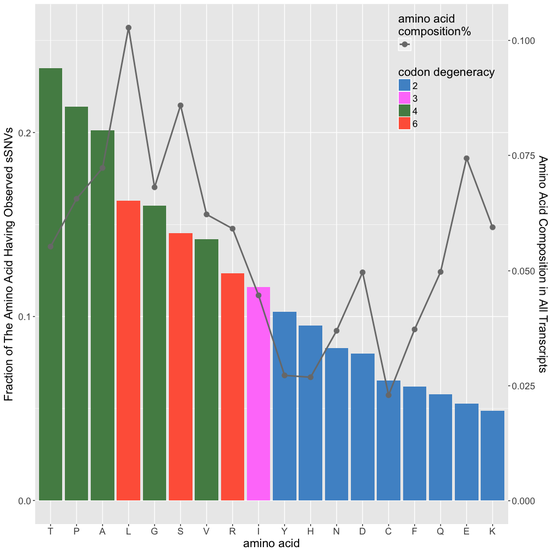
predicting sSNVs effects
machine learning-based classifier to evaluate deleteriousness of synonymous variants
Publications
Check NIH Library for comprehensive list
Contact
- yana@bromberglab.org
- 1510 Clifton Road NE, 1001 O. Wayne Rollins Research Center, Atlanta, GA 30322
- Enter Building, turn right and walk to Room 1001
-
Monday 09:00 to 17:00
Tuesday 09:00 to 17:00
Wednesday 09:00 to 17:00
Thursday 09:00 to 17:00
Friday 09:00 to 17:00 - whereby.com